Diagnosing inefficient biological phosphorus removal
By leveraging LuminUltra’s mail-in GeneCount services, a wastewater treatment plant was able to identify the underlying causes of poor biological phosphorus removal in one of their two bioreactors.
Key takeaway:
An Australian wastewater treatment plant’s operations staff were experiencing inconsistent phosphorus removal across their parallel anaerobic-aerobic bioreactors. By conducting metagenomic analysis, they were able to identify the quantity (cell/mL) and relative abundance of phosphorus accumulating organisms (PAOs) and glycogen accumulating organisms (GAOs).
Learn more below, or contact us for details about LuminUltra’s GeneCount Sample Preservation Kit and Next-Generation Sequencing services
Introduction
A wastewater treatment plant (WWTP) In Australia utilizes enhanced biological phosphorus removal to meet stringent effluent guidelines. The WWTP consists of two parallel anaerobic-aerobic bioreactors (Figure 1). Further phosphorus removal is providing using tertiary coagulation-media filtration.
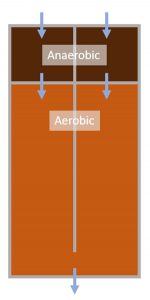
Operations staff were experiencing issues with inconsistent biological phosphorus removal across the two bioreactors. Bioreactor 1 was consistent achieving good biological phosphorus removal, while Bioreactor 2 was not. To investigate why this was occurring, samples of the mixed liquor suspended solids (MLSS) were taken for metagenomic analysis to better understand the microbial community composition.
See also: 4 Guidelines to getting the most of your metagenomic data
Samples were collected from both bioreactors and preserved using LuminUltra’s GeneCount Sample Preservation Kit. This allows DNA to be concentrated and shipped in a small vial at ambient temperature, as opposed to cost prohibitive cold chain transport, all while ensuring data integrity is maintained. Without proper preservation, the microbial community can shift, resulting in inaccurate findings.
In LuminUltra’s DNA sequencing lab, samples were purified then two analyses were run:
- qPCR for Total Prokaryotes – which quantified the total number of prokaryotes (bacteria and archaea) in the sample in cells/mL
- 16S Illumina Sequencing – which identified and quantified (in relative to abundance) all prokaryotes in the sample
The two results were combined to calculate the quantity of each organism identified. The metabolic functions of the organisms were characterized using a literature derived database.
Results and Discussion
The organisms which were of importance to this study were phosphorus accumulating organisms (PAOs) and glycogen accumulating organisms (GAOs). Under specific conditions, PAOs are able to remove phosphorus by accumulating and storing large quantities of polyphosphate. In contrast, CAOs compete with specific PAOs, such as Candidatus Accumulibacter, for vital volatile fatty acids (VFAs), thereby reducing the phosphorus removal efficiency of those organisms.
There were several PAOs and GAOs found in the samples. Results are shown in Table 1.
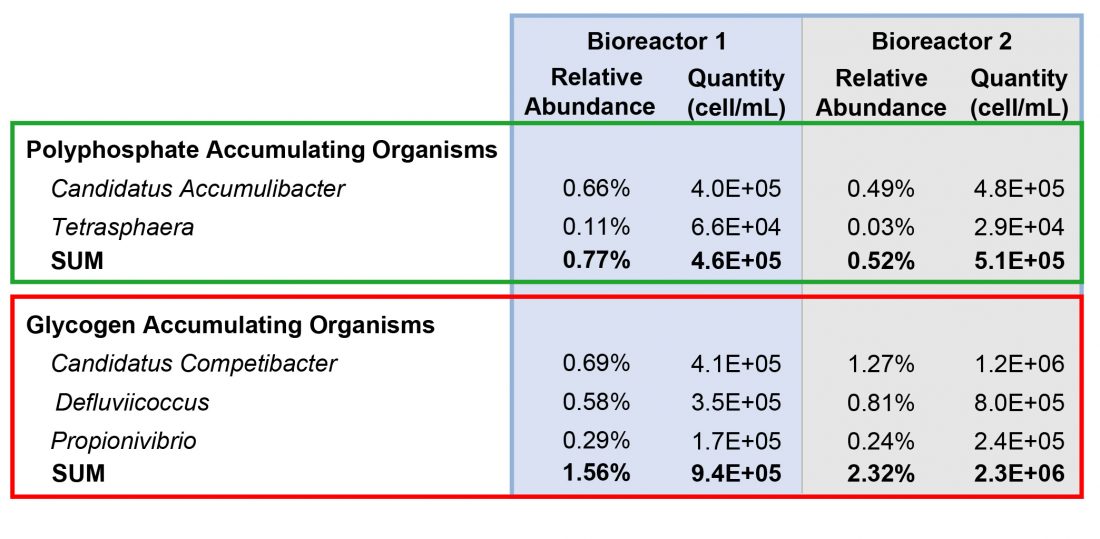
The most abundant PAO was Candidatus Accumulibacter. Bioreactor 1 had a higher relative abundance of both PAOs compared to Bioreactor 2, however the total quantity was similar as Bioreactor 2 had a higher total prokaryote concentration. This highlights the importance of examining the total quantity in addition to the relative abundance.
Bioreactor 2 had a much higher GAO relative abundance and quantity. The quantity in Bioreator 2 was 43% higher than Bioreactor 1. This suggests that GAOs are inhibiting phosphorus removal in Bioreactor 2. GAOs compete with Candidatus Accumulibacter for VFAs, which Candidatus Accumulibacter need to form polyhydroxybutyrate (PHB) storage products. PHB storage products provide energy for Candidatus Accumulibacter in aerobic environments to replicate and form intracellular polyphosphate bonds, thereby removing phosphorus from the wastewater.
Conclusion
Using metagenomic analysis, the underlying cause of poor phosphorus removal in Bioreactor 2 was identified. In general, there was sufficient PAOs, but excessive GAOs which compete for essential VFAs and reduce treatment efficiency. The next steps are to investigate why GAOs proliferated in Bioreactor 2 but not Bioreactor 1. Factors to investigate are pH, influent carbon/phosphorus ratio, solids retention time, VFA type and temperature.