Understanding loss of nitrification at a refinery wastewater treatment plant using 16S rRNA sequencing
What’s inside

This research highlights the utility of advanced biomass characterization in refinery wastewater treatment to understand process upsets and identify shifts within the microbial community that are contributing to changes in treatment performance.
The full version of this article was published in the March 2023 issue of Water Environment & Technology.
Summary
A refinery wastewater treatment plant (WWTP) in the southwestern United States was monitored from June 2017 to November 2020 to investigate nitrogen removal. The refinery started processing additional feedstocks such as light, tight oil-sands, shale oils, and conventional crude oils of various characteristics. The WWTP had initially assumed complete nitrification was occurring as effluent ammonia discharge requirements were continuously met. However, the plant struggled to meet toxicity effluent requirements. This led to the need of immediate investigation, as the plant discharges into protected receiving waters.
16S rRNA gene sequencing and cATP along with other plant process data was routinely collected from bioreactor 1 for the investigation. 16S rRNA gene sequencing was completed using Illumina MiSeq to find the relative abundance of all bacteria and archaea in a sample down to the genus level. cATP is the true quantity of active biomass, the living portion of biomass that can be used to analyse the health and treatment capacity of a system. cATP combined with the relative abundance can be used to calculate the true organism’s concentration.
In September 2019 the WWTP updated the Dissolved Air Floatation (DAF) unit, which was completed by September 2020 due to the operator’s suspicion of insufficient operation. Improvements to the DAF unit included:
- rectifications to the primary oil water separator
- adjusting and repairing the dissolved air flotation separators
- optimizing coagulant and flocculant doses.
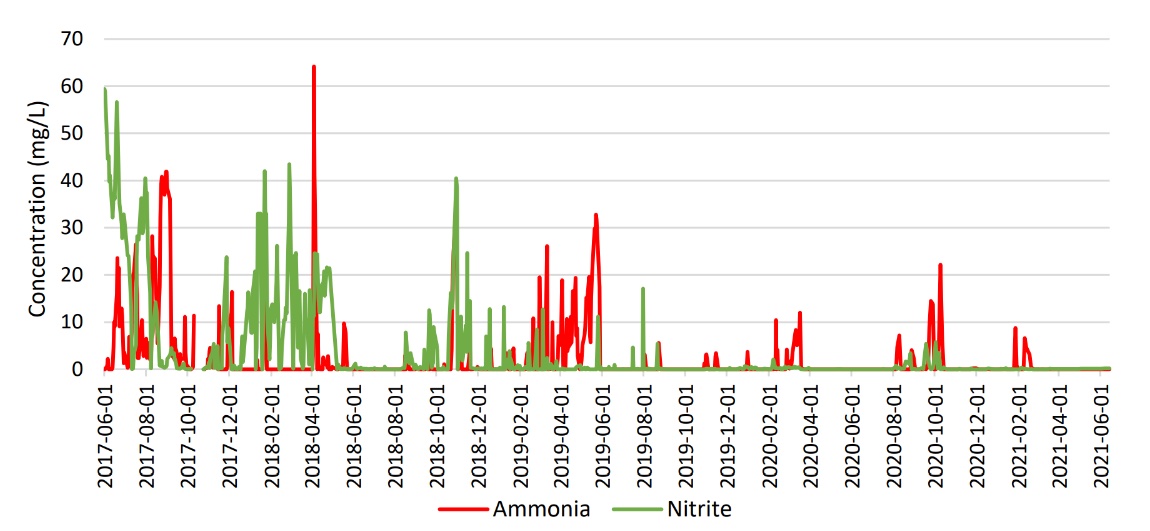
Following September 2019 ammonia and nitrite samples from the bioreactor effluent showed consistent low levels and there were fewer recorded upsets; this can be seen in Figure 1. In February 2020, following the primary treatment improvements, the effluent oil and grease concentrations were consistently lower. During this time, the powdered activated carbon (PAC) — which had been added in 2018 to the bioreactor to assist in dissolved organics removal due to toxicity concerns — was significantly reduced while maintaining equivalent or better treatment efficiency. The system improvements stabilized nitrification and prevented system upsets. Additionally, the updates made to the WWTP proved to show useful insight into the cause and effects to the microbial community.
The four-nitrogen related organism functional groups identified using 16S rRNA gene sequencing are:
- ammonia oxidizing (AOX)
- nitrite oxidizing (NOX)
- denitrifying (DN)
- simultaneous nitrification-denitrification (SND)
The abundance of each functional group was reviewed pre and post primary treatment improvements. The heat map (Figure 2) represents the abundance per group over time: red being low concentration, yellow being moderate concentration, and green being high concentration.
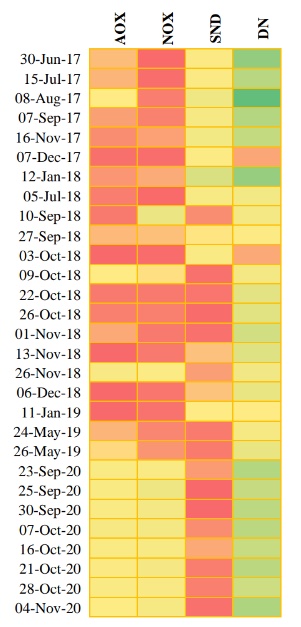
Following the primary treatment improvements, a reduction in SND organisms and an increase in AOX and NOX were observed. The most abundant functional group throughout was DN with Thauera sp. being the most predominant DN taxon. Aerobic, autotrophic Nitrosomonas sp. and Nitrospira sp. were the most predominant genera for AOX and NOX, respectively. DN remained relatively consistent, and the heat map shows following the primary treatment improvements in September 2020 there is consistency and stability within the functional groups. Prior to plant improvements when Thauera concentrations were high, it was observed that there was nitrification loss. Following the improvements, it was observed when Thauera concentrations were high, there were no nitrification losses. In this situation it can be concluded that high DN organisms paired with high SND organisms were not ideal for efficient nitrification. Furthermore, moderate AOX and NOX paired with stable DN are favourable for complete nitrification.
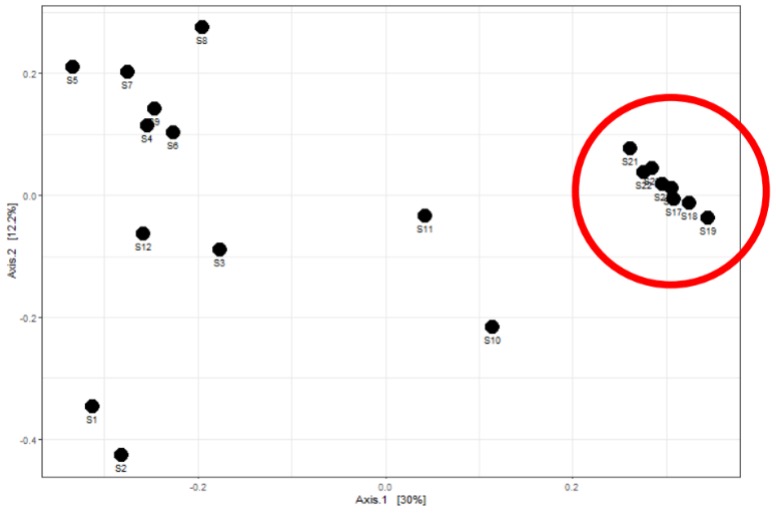
To further investigate the microbial community generated by 16S rRNA gene sequencing, a Principle Coordinate Analysis (PCoA) plot was completed. A PCoA plot can show similarities between a community: samples clustered together represents similarities while samples distancing from one another represents dissimilarities. It was found that following September 2020, after the process improvements, samples clustered together. This can be seen in the enclosed red circle (Figure 3). Prior to September 2020, samples did not cluster together, showing more dissimilarity. The plot shows the improvement in process efficiency, allowing for stable growth and maintenance of desired species.
Active biomass ratio (ABR) represents the percentage of total suspended solids (TSS) that are living microorganisms. It was observed that prior to the WWTP improvements, ABR baseline was below 10% and following the improvements ABR baseline was increased to a 15% to 20% range. This indicates and further proves process improvement as ABR is calculated using active volatile suspended solids (AVSS). AVSS is a direct indication of living biomass as it is calculated using cATP.
By stabilizing desired microbial communities, consistent complete nitrification could be achieved. This was ultimately associated to improvements made to the plant, but able to be verified and explained using cATP and genomic analysis when traditional analytics like microscopy fell short. Routine genomic sequencing and cATP monitoring proved beneficial throughout all ranges of process efficiency.